PLEK2 in head and neck squamous cell carcinoma
Wang J, Sun Z, Wang J, Tian Q, Huang R, Wang H, Wang X, & Han F. Expression and prognostic potential of PLEK2 in head and neck squamous cell carcinoma based on bioinformatics analysis. Cancer Medicine (2021)
How they used Xena
They downloaded data from Xena and also included our Visual Spreadsheet in Figure 12 and our Kaplan Meier plot in Figure 5.
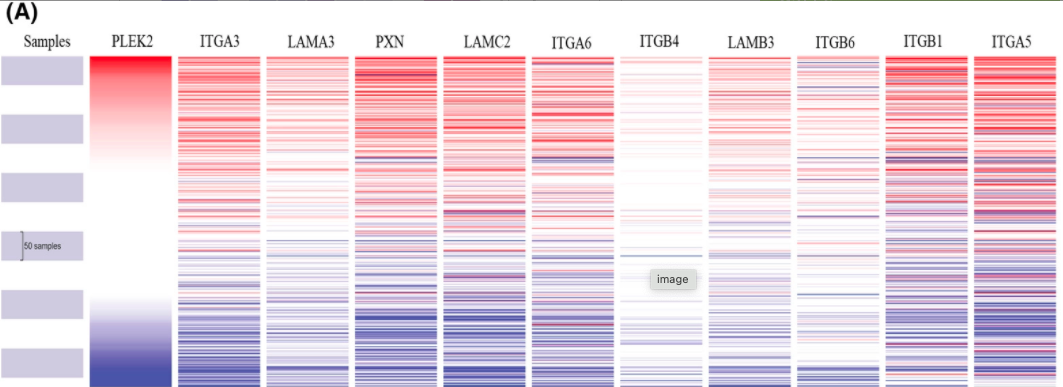
Figure 12A
The comprehensive analysis of ITGA3. The clustering heatmap for hub genes in TCGA HNSCC dataset using the UCSC Xena database.
Join the conversation on Twitter about this publication
Paper
Background
PLEK2 (pleckstrin) could bind to membrane-bound phosphatidylinositols and further promote cell spread. Recently, several studies have noted the importance of PLEK2 in tumor metastasis. However, the role of PLEK2 in head and neck squamous cell carcinoma (HNSCC) remains to be elucidated.
Methods
The PLEK2 expression in HNSCC was identified using Oncomine, Gene Expression Omnibus (GEO), UALCAN databases, and western blot analysis. Prognosis analysis was performed using Kaplan–Meier plotter, DriverDBv3, UALCAN, UCSC Xena, and GEO databases. Single-cell functional analysis was further performed using the cancerSEA database. The PLEK2-related co‑expressed genes were identified, and gene set enrichment analysis was performed using LinkedOmics. Furthermore, the top 10 hub genes were identified using the cytoHubba plug-in of Cytoscape. Then, gene enrichment analysis, pathway activity, and drug sensitivity analyses of the hub genes were performed using the R package “clusterProfiler” and GSCAlite. Finally, the UCSC Xena browser was utilized to explore the hub gene most likely to play a synergic role with PLEK2 in HNSCC.
Results
Elevated expression of PLEK2 was observed in HNSCC and even in HNSCC subgroups based on diverse clinicopathological features, portending a poor prognosis in HNSCC. PLEK2 was correlated with metastasis and hypoxia in HNSCC, and the PLEK2-related co-expressed genes were mainly involved in the focal adhesion pathway. The top 10 hub genes were primarily enriched in focal adhesion, HPV infection, ECM-receptor interaction, and PI3K-AKT signaling pathway, and epithelial–mesenchymal transition pathway was activated. Furthermore, the expression levels of the hub genes were associated with sensitivity and resistance to various small molecules and anti-cancer drugs. Further study suggested that ITGA3 and PLEK2 might be viewed as inextricably linked in facilitating HNSCC metastasis.
Conclusions
In general, PLEK2 might serve as a potential biomarker for the diagnosis of HNSCC and guide the development of targeted therapies for HNSCC.