Investigations into GPC3 in breast cancer
Barman UD, Saha SK, Kader MA, Mostofa Jamal MAH, Sharma SP, Samad A, & Rahman MS. Clinicopathological and prognostic significance of GPC3 in human breast cancer and its 3D structure prediction. Network Modeling Analysis in Health Informatics and Bioinformatics (2020)
How they used Xena
In addition to downloading data from Xena for secondary analysis, they also used Xena to help make Figure 7, where they explored the correlation between GPC3 and RASSF6 mRNA expression.
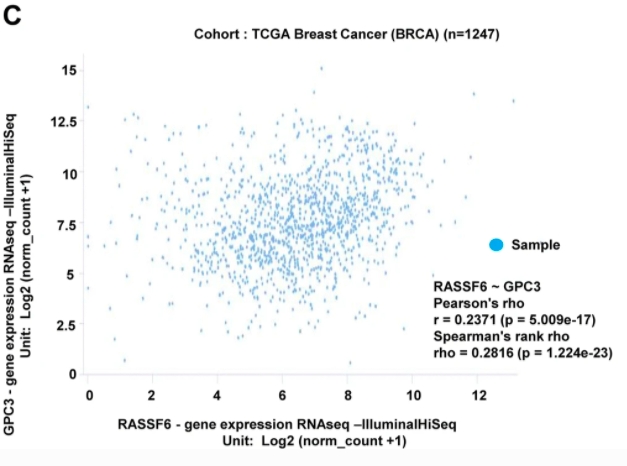
Figure 7 from the paper
Paper
Glypican-3 (GPC3) is a heparan sulfate proteoglycan that may function as a tumor suppressor in breast cancer (BC). To evaluate the prognostic value of GPC3 in BC, systematic analysis was performed in this study. To evaluate gene alteration during breast carcinogenesis, GPC3 expression was analyzed using the Oncomine, GENT, UALCAN, bcGenExMiner, and UCSC Xena databases. The prognostic role of GPC3 in BC was investigated using KM Plotter and PrognoScan databases. Promoter methylation status and heat map of GPC3 were determined using UALCAN and UCSC Xena. GPC3 expression was significantly downregulated in BC compared to that in normal tissues and was correlated with prognosis. However, estrogen receptor and progesterone receptor status were positively correlated with GPC3 expression, whereas basal-like status, triple-negative breast cancer status, and Scarff, Bloom, and Richardson grade criteria were negatively correlated with GPC3 expression. Further analysis indicated that GPC3 was correlated with Ras-association domain family 6 (RASSF6) expression in BC tissues. GPC3 may thus be considered a significant marker for predicting BC prognosis along with RASSF6. Comparative protein modeling of GPC3 was performed using a FASTA formatted sequence from NCBI, in Swiss model, and GPC3 was visualized using PyMOL tools. The GPC3 active site was identified with CASTp server. The Ramachandran plot in PROCHECK showed significant scores for the protein model in its most favored regions. ProSA server indicated the high accuracy of the protein model. However, large-scale and comprehensive studies are needed to clarify these results.